- Research
- Open access
- Published:
Diesel exhaust particle inhalation in conjunction with high-fat diet consumption alters the expression of pulmonary SARS-COV-2 infection pathways, which is mitigated by probiotic treatment in C57BL/6 male mice
Particle and Fibre Toxicology volume 21, Article number: 40 (2024)
Abstract
Background
Both exposure to air pollutants and obesity are associated with increased incidence and severity of COVID-19 infection; however, the mechanistic pathways involved are not well-characterized. After being primed by the transmembrane protease serine 2 (TMPRSS2) or furin protease, SARS-CoV-2 uses the angiotensin-converting enzyme (ACE)-2 receptor to enter respiratory epithelial cells. The androgen receptor (AR) is known to regulate both TMPRSS2 and ACE2 expression, and neuropilin-1 (NRP1) is a proposed coreceptor for SARS-CoV-2; thus, altered expression of these factors may promote susceptibility to infection. As such, this study investigated the hypothesis that inhalational exposure to traffic-generated particulate matter (diesel exhaust particulate; DEP) increases the expression of those pathways that mediate SARS-CoV-2 infection and susceptibility, which is exacerbated by the consumption of a high-fat (HF) diet.
Methods
Four- to six-week-old male C57BL/6 mice fed either regular chow or a HF diet (HF, 45% kcal from fat) were randomly assigned to be exposed via oropharyngeal aspiration to 35 µg DEP suspended in 35 µl 0.9% sterile saline or sterile saline only (control) twice a week for 30 days. Furthermore, as previous studies have shown that probiotic treatment can protect against exposure-related inflammatory outcomes in the lungs, a subset of study animals fed a HF diet were concurrently treated with 0.3 g/day Winclove Ecologic® Barrier probiotics in their drinking water throughout the study.
Results
Our results revealed that the expression of ACE2 protein increased with DEP exposure and that TMPRSS2, AR, NRP1, and furin protein expression increased with DEP exposure in conjunction with a HF diet. These DEP ± HF diet-mediated increases in expression were mitigated with probiotic treatment.
Conclusion
These findings suggest that inhalational exposure to air pollutants in conjunction with the consumption of a HF diet contributes to a more susceptible lung environment to SARS-CoV-2 infection and that probiotic treatment could be beneficial as a preventative measure.
Background
Numerous studies have shown a positive correlation between exposure to traffic-generated air pollution and the progression of respiratory diseases [1,2,3,4,5,6]. The severe acute respiratory syndrome (SARS) novel coronavirus (SARS-CoV-2) that causes the respiratory coronavirus disease (COVID-19) has become endemic, with over 103 million confirmed cases in the United States and nearly 770 million cases worldwide as of 2023 [7, 8]. While air pollution has been linked to increased risk and severity of COVID-19, the mechanisms involved have not yet been fully elucidated [9, 10].
SARS-CoV-2 has been shown to use the angiotensin-converting enzyme 2 (ACE2) receptor for entry into lung epithelial cells [11,12,13]. The spike (S) protein of the coronavirus must be primed by transmembrane protease serine 2 (TMPRSS2) and/or furin protease, which exposes the receptor-binding domain and allows for the fusion of the viral and host membranes [14,15,16,17,18]. The presence of a furin cleavage site is unique to SARS-CoV-2 and is not found in other coronaviruses, which may contribute to the rapid spread of COVID-19. Because of its ability to bind furin-cleaved substrates, neuropilin-1 (NRP1) is a proposed coreceptor for SARS-CoV-2 infection [14,15,16,17].
The androgen receptor (AR) has been demonstrated to regulate TMPRSS2 expression. Activated AR also promotes ACE2 expression, facilitating the priming of coronavirus S proteins and allowing entry into the host cell [18, 19]. This androgen-mediated pathway likely contributes to the observation that men appear to be more susceptible to, as well as have more severe symptoms of, COVID-19 [11,12,13, 18,19,20]. In addition to being the binding site for SARS-CoV-2, ACE2 is essential for normal pulmonary function, with ACE2 dysregulation being reported in several lung diseases, including COPD [11].
Proposed SARS-CoV-2 cellular entrance pathways. Following priming of its spike proteins by the transmembrane protease serine 2 (TMPRSS2) and/or furin protease, SARS-CoV-2 uses the angiotensin-converting enzyme (ACE2) receptor for entry into respiratory epithelial cells. The androgen receptor (AR) has been demonstrated to regulate TMPRSS2 and ACE2 expression, facilitating the priming of coronavirus S proteins and allowing entry into the host cell. Because of its ability to bind furin-cleaved substrates, neuropilin-1 (NRP1) is a proposed coreceptor for SARS-CoV-2 infection. Created withhttp://BioRender.com.
Exposure to air pollution increases susceptibility to various respiratory infections and contributes to inflammatory diseases such as obesity and autoimmune disorders [21, 22]. Studies from China, Italy, and the United States have shown a strong connection between COVID-19 infection and mortality rates and exposure to air pollution [9, 23, 24]. Notably, these regions experience chronic high-level air pollution, so populations most likely have increased underlying levels of pulmonary inflammation compared with regions with better air quality. Although a single air pollutant known to increase susceptibility to COVID-19 has yet to be identified, positive correlations between COVID-19 infection and environmental particulate matter and gases such as O3 and NO2 have been reported [9]. Previous studies by our lab have shown that traffic-generated air pollutants are associated with increased expression of other members of the renin-angiotensin signaling pathway, in which ACE2 plays a key role [25].
The consumption of a high fat (HF)-diet, diet-induced inflammation, and, by extension, obesity have also been identified as risk factors for greater COVID-19 incidence and severity [9, 23, 24, 26]. Additionally, the consumption of foods that score higher on the Dietary Inflammatory Index, many of which are HF, has been shown to be correlated with increased COVID-19 incidence and severity [26]. It has been proposed that obesity causes alterations in the expression and activity of factors associated with SARS-CoV-2 infection. Al Heialy et al. reported that ACE2 and TMPRSS2 are upregulated in obese individuals due to dysregulation of lipid synthesis and clearance, which is characteristic of obesity. The resulting increase in ACE2 expression in the lungs may promote increased susceptibility of viral infection, as well as increased inflammation and exacerbated disease outcomes [27].
Another risk factor contributing to the exacerbation of viral infections is microbiome dysbiosis [27, 29, 30]. The majority of bacteria in the lung microbiome belong to four primary phyla: Firmicutes, Bacteroidetes, Proteobacteria , and Actinobacteria [28]. In healthy individuals, these bacteria typically enter the lungs by aspiration of saliva droplets and are eliminated by coughing, mucociliary clearance, and the local immune response [29]. This cycle of entry and elimination results in a healthy balance of the lung microbiome among the four phyla. This balance is disturbed in lung disease, resulting in an observable shift in the lung microbiome [28,29,30]. An increase in mucus production is characteristic of many pulmonary diseases, allowing for the proliferation of mucus-metabolizing bacteria, especially those belonging to the Proteobacteria phylum [30]. We have previously reported that the consumption of a HF diet and exposure to diesel exhaust particulate matter (DEP) alter the lung microbiome and increase baseline pulmonary inflammation in a wild-type mouse model [31]. However, treatment with probiotics has been shown to diminish Proteobacteria expansion and inflammation in the lungs of these animals [31]. Because of their known immunomodulatory effects, dietary probiotics have been proposed as supplementary treatments for COVID-19 [32].
To further characterize the pathways by which exposure to environmental traffic-generated air pollutants may contribute to susceptibility to COVID-19 infection, we investigated the hypothesis that exposure to DEP increases the expression of factors that mediate the entry of SARS-CoV-2 into pulmonary epithelial cells. Furthermore, we characterized whether exposure to DEP combined with a HF diet further exacerbates these outcomes. Finally, we investigated whether probiotic treatment mitigates DEP- ± HF diet-driven alterations in our exposure model.
Results
Exposure to DEP results in increased ACE2 protein expression in the bronchioles of C57BL/6 mice
To investigate whether subchronic inhalational exposure to DEP and consumption of a HF diet alter ACE2 expression, we measured ACE2 protein expression by immunofluorescent histology (Fig. 1A-M) and mRNA transcript expression by RT-qPCR (Fig. 1N). We observed a significant increase in ACE2 protein expression in experimental groups that were exposed to DEP, regardless of diet. Compared with that in the LF + CON group, ACE2 protein expression was significantly greater in the LF + DEP (p < 0.0001) and HF + DEP groups (p < 0.0001). Compared with that in the HF + CON group, there was a significant increase in ACE2 protein expression in the LF + DEP (p < 0.0001) and HF + DEP groups (p < 0.0001). There was no significant difference in protein expression observed among the exposure groups. Exposure F = 71.07, diet F = 2.494, and exposure x diet F = 0.08277. There was no significant difference in ACE2 mRNA transcript expression between the groups.
Exposure to DEP results in increased angiotensin-converting enzyme 2 (ACE2) receptor protein expression in the bronchioles of C57BL/6 mice. Representative images of ACE2 expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates ACE2 expression, and blue fluorescence indicates nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung ACE2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of ACE2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA. The brightness of the nuclei image panels was increased by 40% and the overlay images were increased by 10%, as shown in this figure
Probiotic treatment mitigated the DEP-mediated increase in ACE2 receptor protein expression in the lungs of C57BL/6 mice
To determine whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in ACE2 expression, we measured ACE2 protein expression via immunofluorescent histology (Fig. 2A-M) and mRNA transcript expression by RT-qPCR (Fig. 2N). We observed a significant decrease in ACE2 protein expression in the HF + CON + PRO (p < 0.0001) and HF + DEP + PRO (p < 0.0001) groups compared with the HF + DEP group. There was no significant difference in ACE2 protein expression between the probiotic groups and the HF + CON group. Exposure F = 43.91, probiotics F = 38.18, exposure x probiotics F = 46.77.
We also observed a significant decrease in ACE2 mRNA expression in the HF + CON + PRO (p = 0.0012) and HF + DEP + PRO (p = 0.0017) groups compared to the HF + CON group. Similarly, ACE2 mRNA expression was significantly lower in the HF + CON + PRO (p = 0.0003) and HF + DEP + PRO (p = 0.0005) groups than in the HF + DEP group. There was no significant difference in ACE2 mRNA expression between the HF + CON and HF + DEP groups or between the probiotic treated groups. Exposure F = 32.10, probiotics F = 0.5418, exposure x probiotics F = 0.4563.
Probiotic treatment mitigates the DEP-mediated increase of angiotensin-converting enzyme 2 (ACE2) protein expression in the bronchioles of C57BL/6 mice. Representative images of ACE2 expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates ACE2 expression, the blue fluorescence indicates nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung ACE2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of ACE2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA. The brightness of the nuclei image panels was increased by 40% and the overlay images were increased by 10%, as shown in this figure
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased TMPRSS2 expression in the lungs of C57BL/6 mice
To assess whether subchronic inhalational exposure to DEP and consumption of a HF diet alter TMPRSS2 expression, we measured TMPRSS2 protein expression by immunofluorescent histology (Fig. 3A-M) and mRNA transcript expression by RT-qPCR (Fig. 3N). We observed a significant increase in TMPRSS2 protein expression in the HF + DEP group compared to the LF + CON (p = 0.0004), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. Compared with the LF + CON group, the LF + DEP (p < 0.0001) and HF + CON (p = 0.0004) groups presented significantly lowerTMPRSS2 protein expression. Compared to the LF + DEP group, the HF + CON (p = 0.0273) group displayed significantly increased TMPRSS2 protein expression was significantly increased. Exposure F = 1.325, diet F = 16.98, exposure x diet F = 56.07.
TMPRSS2 mRNA expression was not significantly different between the LF + CON, LF + DEP, and HF + CON groups, but the expression of TMPRSS2 mRNA in the HF + DEP group was significantly greater than that in the LF + DEP (p = 0.0016) and HF + CON (p = 0.0067) groups. Exposure F = 0.7838, diet F = 2.562, exposure x diet F = 12.77.
Exposure to DEP in conjunction with consumption of a HF diet results in increased transmembrane serine protease 2 (TMPRSS2) expression in the bronchioles of C57BL/6 mice. Representative images of TMPRSS2 expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or fed a LF (D-F) or HF diet (J-L) and exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates TMPRSS2 expression, and blue fluorescence indicates nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung TMPRSS2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of TMPRSS2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM, with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared with HF + CON by two-way ANOVA
Probiotic treatment mitigated the DEP- and HF diet-mediated increase of TMPRSS2 protein expression in the lungs of C57BL/6 mice
To establish whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in TMPRSS2 expression, we measured TMPRSS2 expression by immunofluorescent histology (Fig. 4A-M) and mRNA transcript expression by RT-qPCR (Fig. 4N). We observed a significant decrease in TMPRSS2 protein expression in groups treated with probiotics (HF + CON + PRO (< 0.0001), HF + DEP + PRO (< 0.0001)) compared to the HF + DEP group. Within the probiotic groups, TMPRSS2 protein expression was observed to be significantly decreased in the HF + DEP + PRO (p = 0.0049) group compared to the HF + CON + PRO group. Exposure F = 20.93, probiotics F = 3.547, exposure x probiotics F = 38.84.
TMPRSS2 mRNA expression was not found to be statistically different between the HF + CON, HF + DEP, and HF + DEP + PRO groups. TMPRSS2 mRNA expression in the HF + CON + PRO (p = 0.0275) group was observed to be significantly lower than that of the HF + DEP group. Exposure F = 2.137, probiotics F = 4.328, exposure x probiotics F = 0.1592.
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in transmembrane serine protease 2 (TMPRSS2) protein expression in the bronchioles of C57BL/6 mice. Representative images of TMPRSS2 expression in the bronchioles of C57Bl/6 mice fed a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates TMPRSS2 expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung TMPRSS2 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of TMPRSS2 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Exposure to diesel exhaust particulate matter in conjunction with the consumption of a HF diet results in increased androgen receptor protein expression in the bronchioles of C57BL/6 mice
To determine if subchronic inhalational exposure to DEP and consumption of a HF diet alters AR expression, we measured AR protein expression by immunofluorescent histology (Fig. 5A-M) and mRNA transcript expression by RT-qPCR (Fig. 5N). We observed a significant increase in AR protein expression in the HF + DEP group compared with the LF + CON (p < 0.0001), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. There was no significant difference observed between the LF + CON, LF + DEP, and HF + CON groups. Exposure F = 10.21, diet F = 17.17, exposure x diet F = 22.58.
AR mRNA expression was significantly greater in the LF + DEP (p = 0.0188) group than in the LF + CON group as well as decreased in the HF + DEP group (p = 0.0307) compared to the LF + DEP group. Exposure F = 1.669, diet F = 1.270, exposure x diet F = 5.069.
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased androgen receptor (AR) protein expression in the bronchioles of C57BL/6 mice. Representative images of AR expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates AR expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung AR mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of AR mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in androgen receptor protein expression in the bronchioles of C57BL/6 mice
To ascertain whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in AR expression, we measured AR expression via immunofluorescent histology (Fig. 6A-M) and mRNA transcript expression by RT-qPCR (Fig. 6N). We observed a significant decrease in AR protein expression in the groups treated with probiotics [HF + CON + PRO (p < 0.0001), HF + DEP + PRO (p < 0.0001)] compared with the HF + DEP group. There was no significant difference observed between the HF + CON and probiotic groups. Exposure F = 14.03, probiotics F = 5.946, exposure x probiotics F = 28.60. There was no significant difference in AR mRNA expression observed between groups.
Probiotic treatment mitigated the DEP- and HF diet-mediated increase of androgen receptor (AR) protein expression in the bronchioles of C57BL/6 mice. Representative images of AR expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates AR expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung AR mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of AR mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Exposure to diesel exhaust particulate matter in conjunction with the consumption of a HF diet results in increased NRP1 protein expression in the bronchioles of C57BL/6 mice
To investigate whether subchronic inhalational exposure to DEP and consumption of a HF diet alter NRP1 expression, we measured NRP1 protein expression via immunofluorescent histology (Fig. 7A-M) and mRNA transcript expression by RT-qPCR (Fig. 7N). We observed a significant increase in NRP1 protein expression in the HF + DEP group compared with the LF + CON (p < 0.0001), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. There was no significant difference observed between the two LF groups. NRP1 protein expression was decreased in the HF + CON group compared with the two LF groups [LF + CON (p = 0.0159), LF + DEP (p < 0.0001)]. Exposure F = 44.21, diet F = 2.039, exposure x diet F = 27.47.
NRP1 mRNA expression was observed to be significantly greater in the LF + DEP group than in the LF + CON (p = 0.0205), HF + CON (p = 0.0125), and HF + DEP (p = 0.0239) groups. There was no significant difference in NRP1 mRNA expression among the LF + CON, HF + CON, and HF + DEP groups. Exposure F = 3.732, diet F = 3.976, exposure x diet F = 2.236.
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased neuropilin 1 (NRP1) protein expression in the bronchioles of C57BL/6 mice. Representative images of NRP1 expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates NRP1 expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung NRP1 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of NRP1 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA
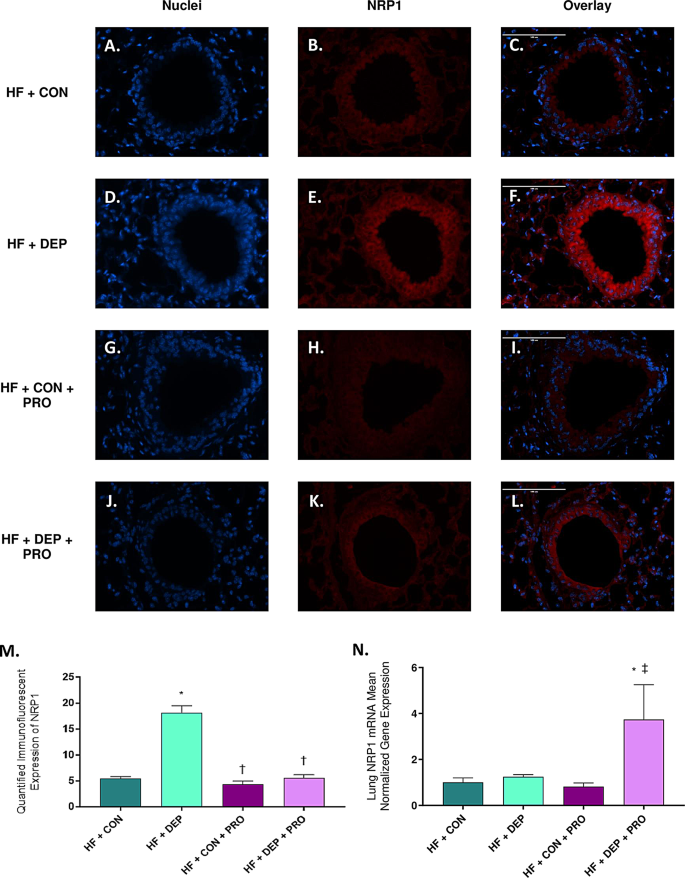
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in NRP1 protein expression in the bronchioles of C57BL/6 mice
To determine whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in NRP1 expression, we measured NRP1 expression via immunofluorescent histology (Fig. 8A-M) and mRNA transcript expression by RT-qPCR (Fig. 8N). We observed a significant decrease in NRP1 protein expression in groups treated with probiotics [HF + CON + PRO (p < 0.0001), HF + DEP + PRO (p < 0.0001)] compared with the HF + DEP group. There was no significant difference observed between the HF + CON and probiotic groups. Exposure F = 38.85, probiotics F = 41.20, exposure x probiotics F = 27.18.
We also observed a significant increase in NRP1 mRNA expression in the HF + DEP + PRO group compared with the HF + CON (p = 0.0345) and HF + CON + PRO (p = 0.0161) groups. There was no significant difference in NRP1 mRNA expression among the HF + CON, HF + DEP, and HF + CON + PRO groups. Exposure F = 1.792, probiotics F = 3.385, exposure x probiotics F = 2.405.
Probiotic treatment mitigates DEP- and HF diet-mediated increase of neuropilin 1 (NRP1) protein expression in the bronchioles of C57BL/6 mice. Representative images of NRP1 expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates NRP1 expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung NRP1 mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of NRP1 mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased furin protein expression in the bronchioles of C57BL/6 mice
To establish whether subchronic inhalational exposure to DEP and consumption of a HF diet alter furin expression, we measured furin expression via immunofluorescent histology (Fig. 9A-M) and mRNA transcript expression by RT-qPCR (Fig. 9N). We observed a significant increase in furin protein expression in the HF + DEP group compared with the LF + CON (p < 0.0001), LF + DEP (p < 0.0001), and HF + CON (p < 0.0001) groups. There was no significant difference observed between the LF + CON, LF + DEP, and HF + CON groups. Exposure F = 30.95, diet F = 28.62, and exposure x diet F = 45.02. We also observed no significant difference in furin mRNA expression among the four groups.
Exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet results in increased furin protein expression in the bronchioles of C57BL/6 mice. Representative images of furin expression in the bronchioles of C57Bl/6 mice on a control (LF; A-C) or high-fat (HF; G-I) diet exposed to saline (CON) or on a LF (D-F) or HF diet (J-L) exposed to diesel exhaust particles (DEP – 35 µg PM) twice a week for a total of 30 days. Red fluorescence indicates furin expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung furin mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of furin mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification; scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to LF + CON, †p < 0.05 compared to LF + DEP, ‡p < 0.05 compared to HF + CON by two-way ANOVA
Probiotic treatment mitigated the DEP- and HF diet-mediated increase in furin protein expression in the bronchioles of C57BL/6 mice
To determine whether probiotic treatment mitigates DEP- ± HF diet-mediated alterations in furin expression, we measured furin expression by immunofluorescent histology (Fig. 10A-M) and mRNA transcript expression by RT-qPCR (Fig. 10N). We observed a significant decrease in furin protein expression in the groups treated with probiotics [HF + CON + PRO (p < 0.0001), HF + DEP + PRO (p < 0.0001)] compared with the HF + DEP group. Furin protein expression was significantly lower in the HF + CON + PRO group than in the HF + CON (0.0360) and HF + DEP + PRO (p = 0.0077) groups. Exposure F = 46.02, probiotics F = 60.13, exposure x probiotics F = 12.00.
Furin mRNA expression was decreased in the HF + CON + PRO group compared with the HF + CON (p = 0.0399) and HF + DEP + PRO (p = 0.0022) groups. There was no significant difference observed in furin mRNA expression among the HF + CON, HF + DEP, and HF + DEP + PRO groups. Exposure F = 0.04313, probiotics F = 3.122, exposure x probiotics F = 7.989.
Probiotic treatment mitigates DEP- and HF diet-mediated increase of furin protein expression in the bronchioles of C57BL/6 mice. Representative images of furin expression in the bronchioles of C57Bl/6 mice on a high-fat (HF) diet exposed to either (A-C) saline (CON), (D-F) DEP – 35 µg PM, (G-I) saline and probiotics (PRO) − 0.3 g/day (~ 7.5 × 107 cfu/day), or (J-L) DEP and probiotics. Red fluorescence indicates furin expression, and blue fluorescence indicates the nuclear staining (Hoechst). The right panels (C, F, I, L) are overlayed figures of the left (blue; A, D, G, J) and center (red; B, E, H, K) panels. M Graph of histological analysis of lung furin mean fluorescence (minimum of 4 sections per slide and n = 3 per group) and N mean normalized gene expression of furin mRNA transcript expression within the lungs, as determined by RT-qPCR (n = 6–8 per group). 40x magnification, scale bar = 100 μm. The data are presented as mean ± SEM with *p < 0.05 compared to HF + CON, †p < 0.05 compared to HF + DEP, ‡p < 0.05 compared to HF + CON + PRO by two-way ANOVA
Discussion
The present study demonstrated that subchronic inhalational exposure to diesel exhaust particulate matter in conjunction with consumption of a HF diet contributes to a lung environment that may be more susceptible to SARS-CoV-2 infection, which can be attenuated through oral probiotic treatment. Specifically, our results revealed that the expression of the ACE2 protein increases with DEP exposure and that the expression of pulmonary proteins TMPRSS2, AR, NRP1, and furin increases with DEP exposure in conjunction with the consumption of a HF diet. These DEP- ± HF diet-mediated alterations in expression were mitigated when the lung microbiome was diversified and re-established through probiotic treatment throughout the exposure period.
The observed upregulation of SARS-CoV-2 infection pathways in DEP-exposed mice fed a HF diet al.igns with epidemiological data reporting an increased incidence and severity of COVID-19 in regions with increased air pollution and in obese individuals [9, 23, 24, 27]. While ACE2 is known to be used by SARS-CoV-2 for cellular entry, whether NRP1 is utilized in the same manner has not been confirmed, although strong evidence suggests that this occurs. Previous studies reported increased COVID-19 susceptibility due to obesity-related alterations in ACE2, allowing for increased viral binding potential [26]. In agreement with these reports, our findings indicate that consumption of a HF diet combined with exposure to inhaled DEP causes the upregulation of all of the members of the TMPRSS2/furin and ACE2 SARS-CoV-2 infection pathways. Additionally, our findings indicate that these combined insults of HF diet and inhaled DEP cause further increases in SARS-CoV-2 infection potential due to the observed upregulation of NRP1.
Most of the significant changes we observed were through histological analysis of the bronchioles. These findings suggest that our 30-day DEP and HF diet exposure resulted in local alterations in the bronchiolar epithelium rather than whole-organ lung alterations in the expression of SARS-CoV-2 infection pathways. This is further supported by the fact that our RT-qPCR (Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and 11) and ELISA (Supplementary Figs. 1 and 2) analyses, which were performed with whole lung tissue homogenate, produced largely insignificant or unexpected results compared with our immunofluorescent histological analysis. These local alterations are consistent with our routes of exposure, as the bronchiolar epithelium is the primary location of contact with the inhaled DEP and also orally administered probiotics via aspiration. Because of the observed alterations in SARS-CoV-2 pathways locally at the bronchiolar epithelium in our study, increased susceptibility to other respiratory infections may also be conferred with these exposures. This premise aligns with previous data reporting increased inflammation, immunosuppression, and infection in the lungs with exposure to air pollutants [5, 33].
In addition to observing direct alterations in expression of SARS-CoV-2 infection pathway members in the present study, our laboratory previously reported alterations in the lung microbiome and in lung immunity in the same animals [31]. Specifically, increases in BALF IgG and IgA (indicators of the lung immune response), increased macrophage infiltration, and increased mucus production in response to DEP exposure ± HF diet were observed [31]. TNF-α and ROS/RNS production were also shown to be significantly increased in the lungs of animals exposed to DEP [31]. Additionally, shifts in the lung microbiome composition were recorded, most notably the expansion of the Proteobacteria phylum, which is indicative of increased inflammation and is associated with various lung diseases [31, 34]. These findings suggest that the DEP ± HF diet-mediated alterations in the microbiome and immune response in the lung may contribute to more severe respiratory disease outcomes [31, 34]. There were also notable mitigative and immune-regulating effects of probiotic supplementation, much like those reported in the present study. DEP ± HF diet-mediated increases in Proteobacteria expansion, TNF-α expression, and ROS/RNS production were all attenuated with probiotic-treatment [31]. Our findings of attenuation of the expression of factors that mediate SARS-CoV-2 infection align with these previously reported protective effects of probiotic supplementation in the lungs. Although the underlying mechanism has not yet been defined, it is important to note that the probiotics used were suspended in water, likely resulting in alterations in the lung microbiome via microaspiration [31, 35, 36].
While these previous studies have reported that DEP exposure ± HF diet directly alters the respiratory microbiome and promotes inflammation, there are also likely effects on the gut-lung axis signaling pathway, which may also contribute to local and systemic inflammatory signaling as well as the disruption of gut permeability. LPS, an endotoxin derived from the outer membrane of gram-negative bacteria in the gut, is known to signal through toll-like receptors (TLRs), such as TLR-2 and − 4, in addition to other receptors in the lung, leading to increased ROS production and inflammatory signaling [37, 38]. Our laboratory previously reported that DEP exposure and HF diet significantly increase expression of TLR-2 and TLR-4 receptors in the lung [31] and the level of circulating LPS [35], which is indicative of increased systemic inflammation. In addition to the increase in circulating LPS in DEP-exposed animals, we also observed elevated systemic inflammatory signaling, most notably TNF-α, interleukin (IL)-3, and IL-1α, and pulmonary ROS [31, 35]. Increased LPS is also indicative of increased gut permeability. Previous analyses by our laboratory revealed that probiotic supplementation attenuated the DEP- and HF diet-driven increases in circulating LPS, suggesting that gut integrity decreases with DEP and a HF diet and is restored with probiotics [35]. In this same study, our laboratory also observed the DEP exposure and a HF diet promoted shifts in the abundance of gut Proteobacteria (increased) and Bifidobacteria (decreased), which were mitigated by probiotic supplementation [35]. Bifidobacteria are known to be the main producers of short chain fatty acids (SCFAs), which are key immune system-regulating molecules, whereas Proteobacteria expansion is indicative of increased inflammation. Thus, these observed shifts in the microbiome also confer alterations in immune signaling [34, 39, 40]. The collective findings from this study suggest that DEP exposure and a HF diet not only result in the pulmonary effects reported in the results section, but also alter gut-lung axis signaling, and promote ROS, inflammation, and immune modulation systemically.
While the current findings report novel outcomes related to DEP exposure, HF diet consumption, and increased expression of factors that mediate SARS-CoV-2 susceptibility, some limitations should be noted. First, this study utilized a male wild-type mouse model because of the increased incidence and severity of COVID-19 symptoms in male humans; however, how the SARS-CoV-2 infection pathways analyzed in this study are altered in females is currently unknown. In addition, we only analyzed outcomes in the reported pathways following a 30-day exposure ± HF diet protocol, rather than at multiple timepoints throughout the duration of the exposure period. Notably, the DEP exposure was performed twice a week, which could allow for intermediate clearance of particles between exposures. As detailed previously by our laboratory, while the dose chosen for the current study (approximately 10 µg/mouse/day) was higher than that experienced by typical human environmental exposures, the findings in the lungs are in agreement with those of previous DEP exposure studies [31, 36]. We also recognize that the DEP used in the current study are not necessarily representative of all vehicle-generated PM found in ambient air pollution; however, the toxicological effects of these particular DEP in the lungs have been well characterized in previously reported studies [31]. Finally, the current study focused only on investigating the expression of members of the SARS-CoV-2 infection pathway that involve fusion of the viral and cell membranes, whereas more recent studies suggest that SARS-CoV-2 can also infect cells via endogenous receptor-mediated endocytosis mechanisms [17].
Notably, the experimental design of the current study did not include probiotic treatment in the low-fat diet exposure groups because our original aims focused on characterizing the potential mitigative effects of probiotics on inflammation and microbiome dysbiosis caused by a HF diet + DEP exposure. However, our laboratory has recently completed studies that will examine the effects of probiotics on a DEP-exposed mouse model without the insult of a HF diet, which will be published as results are obtained. On the basis of our previously reported findings [31, 35, 36], we hypothesize that we will observe similar protective effects in both the lungs and gut with probiotic usage in the absence of a HF diet.
Even when considering these limitations, this study provides preliminary insight into the potential underlying mechanism of SARS-CoV-2 susceptibility and infection, which may be exacerbated by exposure to environmental air pollutants and/or the consumption of a HF diet as well as the potential for probiotics to be used as supplemental or preventative treatments.
Conclusions
The relationships among air pollutant exposure, diet, and COVID-19 incidence and severity are not well-understood, nor are the potential benefits of probiotic treatment. Thus, we aimed to study the effects of DEP inhalation, HF diet consumption, and oral probiotic supplementation on the expression of SARS-CoV-2 infection pathways. Our results revealed that the expression of the ACE2 protein is increased with DEP exposure and that the expression of TMPRSS2, AR, NRP1, and furin proteins increased with DEP exposure in conjunction with a HF diet in the bronchioles. The DEP- and/or HF diet-mediated increases in expression were mitigated with probiotic treatment. These findings suggest that inhalational exposure to air pollutants in conjunction with the consumption of a HF diet contributes to a more susceptible lung environment to SARS-CoV-2 infection and that probiotic treatment could potentially be used as an adjunctive or preventative measure.
Materials and methods
Animals and exposures
Eight-week-old C57BL/6 male mice were placed on either a HF (HF) (n = 24) or low-fat control (LF) (n = 24) diet and randomly sorted to be exposed to either 35 µg diesel exhaust particles (DEP; NIST Standard Reference Material #2975), suspended in 35 µl 0.9% sterile saline (n = 12 per diet group) or sterile saline only (control) (n = 12 per diet group) via oropharyngeal aspiration, two times per week for 30 days, as previously described by our laboratory [31, 35, 36].
A separate set of mice (n = 24) were placed on the HF diet, exposed to either DEP (n = 12) or control (n = 12), and treated with a dose of 0.3 g/day (~ 7.5 × 108 cfu/day) of Ecologic® Barrier probiotics (Winclove Probiotics, Amsterdam, Netherlands) in the drinking water over the course of the exposures, as previously described by our lab [31, 35, 36]. The probiotics used includes a blend of the following bacterial strains: Bifidobacterium bifidum W23, Bifidobacterium lactis W51, Bifidobacterium lactis W52, Lactobacillus acidophilus W37, Lactobacillus brevis W63, Lactobacillus casei W56, Lactobacillus salivarius W24, Lactococcus lactis W19 and, Lactococcus lactis W58.
Lung tissues
Within 12 h of the final exposure, the trachea/bronchi/lungs of all of the animals were collected under sterile conditions from all animals (n = 8/grp). The left lung was ligated, removed from the right lung, and snap frozen. The right lung was flushed with sterile, cold PBS and the BALF was collected, then snap frozen. A subset of the lungs (n = 4–5) was flushed with 10% NBF under pressure and embedded for tissue staining.
Immunofluorescent histology
For immunofluorescence analyses, lungs that were embedded in paraffin and sectioned at 5 μm thickness were processed and single immunofluorescence was used to detect and localize expression of the ACE2, TMPRSS2, AR NRP1, and furin.
The sectioned lung tissues were deparaffinized with HistoChoice Clearing Agent three times for 5 min each time at room temperature, rehydrated, and stained via methods previously described by our laboratory with the following alterations to the antigen retrieval procedure: EDTA (pH = 8) in place of citrate buffer, heating for 5 min at 20% power instead of 30 s at full power, and an additional set of three 5-minute rinses in 1X PBS following heating [21, 25, 41,42,43]. The primary antibodies used were against ACE2 (1:250; Santa Cruz Biotechnology, sc-390851), TMPRSS2 (1:500; Proteintech, 14437-1-AP), AR (1:500; Proteintech, 22089-1-AP), furin (1:500; Santa Cruz Biotechnology, sc-133142), and NRP1 (1:500; Santa Cruz Biotechnology, sc-5307). The secondary antibodies used were anti-mouse Alexa Fluor 546 for ACE2, furin, and NRP1 staining and anti-rabbit Alexa Fluor 555 for both TMPRSS2 and AR staining.
Slides were imaged using fluorescent microscopy at 20x and 40x with the appropriate excitation/emission filter, digitally recorded, RGB overlay signals were split and analyzed for mean fluorescence using image densitometry with ImageJ software (NIH). A minimum of 4 sections per slide and n = 2–3 per group, were used for analysis. 40x images were used for image quantification.
RT-qPCR
RNA was isolated from lung tissue and (n = 6–8 per group) using an RNAEasy Mini kit (Qiagen, Valencia, CA) per kit instructions and cDNA was synthesized using an iScript cDNA Synthesis kit (Biorad, Cat. #170891). RT-qPCR was done to confirm the expression of ACE2, TMPRSS2, AR, NRP1, and furin using the appropriate primers (Table 1) and PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Cat. #A25742). Samples were run on a Biorad CFX96 and analyzed using Biorad CFX Manager software as previously described by our laboratory [21, 43,44,45,46].
Lung protein extraction
A total of 20–40 mg of lung tissue was placed in lysis buffer (297 µL of RIPA buffer and 3 µL protease inhibitor cocktail (Calbiochem, Millipore Sigma, St. Louis, MO, cat. #535140) and homogenized in bead beater for 15 min then centrifuged at 15,000 rpm for 5 min at 4 °C. The supernatant was removed and the mixture was sonicated for 15 min then centrifuged again. The supernatant was removed and frozen at -20 °C. The samples were checked for purity and protein quantification using a Take3™ Micro-Volume Plate (BioTek, Winooski, VT) read on Cytation™5 Imaging Reader with Gen5 Software (BioTek, Winooski, VT).
ELISA
TMPRSS2 concentration was measured using protein extracted from lung tissues (n = 6 per group) following the manufacturer’s protocol (Biomatik, #50-228-6433). ELISA was read on a Cytation5 plate reader at 450 nm absorbance. The samples were processed in duplicates and values were determined from a known value standard curve using a second-order polynomial (quadratic) curve fit.
Statistical analysis
A two-way ANOVA with a post hoc Holm-Sidak multiple comparison all-pairwise test was used to analyze data across groups of diet (variable 1) vs. exposure (variable 2)-dependent outcomes, or interactions of diet x exposure, as well as across groups of exposure vs. probiotic usage (variable 3), or interactions of exposure x probiotic usage. Statistical analyses were performed with GraphPad Prism 10. A p < 0.05 was considered statistically significant.
Data availability
Some data is provided within the manuscript or supplementary information files. All remaining data will be made available from the corresponding author upon reasonable request.
Abbreviations
- ACE2:
-
Angiotensin-converting enzyme 2
- AR:
-
Androgen receptor
- BALF:
-
Bronchoalveolar lavage fluid
- CON:
-
Control (saline)
- COPD:
-
Chronic obstructive pulmonary disorder
- COVID-19:
-
Coronavirus Disease 2019
- DEP:
-
Diesel exhaust particulate
- HF:
-
HF
- IgA:
-
Immunoglobulin A
- IgG:
-
Immunoglobulin G
- LF:
-
Low-fat
- NRP1:
-
Neuropilin 1
- PM:
-
Particulate matter
- PRO:
-
Probiotic
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- TMPRSS2:
-
Transmembrane serine protease 2
- TNF-α:
-
Tumor necrosis factor-alpha
References
Burnett T, Brook R, Dann J, Delocla T, Philips C, Cakmak O. Association between particulate- and Gas-Phase Components of Urban Air Pollution and Daily Mortality in eight Canadian cities. Inhal Toxicol. 2000;12(sup4):15–39.
Goldberg MS, Burnett RT, Bailar JC, Tamblyn R, Ernst P, Flegel K, et al. Identification of persons with cardiorespiratory conditions who are at risk of dying from the acute effects of ambient air particles. Environ Health Perspect. 2001;109(suppl 4):487–94.
Kelly FJ, Fussell JC. Air pollution and airway disease: air pollution and airway disease. Clin Exp Allergy. 2011;41(8):1059–71.
Faustini A, Stafoggia M, Colais P, Berti G, Bisanti L, Cadum E, et al. Air pollution and multiple acute respiratory outcomes. Eur Respir J. 2013;42(2):304–13.
Andersen ZJ, Hvidberg M, Jensen SS, Ketzel M, Loft S, Sørensen M, et al. Chronic obstructive Pulmonary Disease and Long-Term exposure to traffic-related Air Pollution: a Cohort Study. Am J Respir Crit Care Med. 2011;183(4):455–61.
Karakatsani A, Analitis A, Perifanou D, Ayres JG, Harrison RM, Kotronarou A, et al. Particulate matter air pollution and respiratory symptoms in individuals having either asthma or chronic obstructive pulmonary disease: a European multicentre panel study. Environ Health. 2012;11(1):75.
CDC. Centers for Disease Control and Prevention. 2020 [cited 2023 Aug 3]. COVID Data Tracker. https://covid.cdc.gov/covid-data-tracker
WHO Coronavirus (COVID-19.) Dashboard [Internet]. [cited 2023 Aug 3]. https://covid19.who.int
Zhu Y, Xie J, Huang F, Cao L. Association between short-term exposure to air pollution and COVID-19 infection: evidence from China. Sci Total Environ. 2020;727:138704.
Martelletti L, Martelletti P. Air Pollution and the Novel Covid-19 Disease: a putative disease risk factor. SN Compr Clin Med. 2020;2(4):383–7.
Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and Regulator of the renin-angiotensin system: celebrating the 20th anniversary of the Discovery of ACE2. Circ Res. 2020;126(10):1456–74.
Kreutz R, Algharably EAEH, Azizi M, Dobrowolski P, Guzik T, Januszewicz A, et al. Hypertension, the renin–angiotensin system, and the risk of lower respiratory tract infections and lung injury: implications for COVID-19. Cardiovasc Res. 2020;116(10):1688–99.
Wiener RS, Cao YX, Hinds A, Ramirez MI, Williams MC. Angiotensin converting enzyme 2 is primarily epithelial and is developmentally regulated in the mouse lung. J Cell Biochem. 2007;101(5):1278–91.
Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–60.
Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–5.
Berkowitz RL, Ostrov DA. The elusive coreceptors for the SARS-CoV-2 spike protein. Viruses. 2023;15(1):67.
larissa.menescal. PromoCell. 2021 [cited 2023 Jul 26]. Furin & Cathepsin L. New targets for Covid-19 drug screening. https://promocell.com/news/new-targets-for-covid-19-drug-screening-sars-inhibitors-sars-research-covid-research/
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–e2808.
Mohamed MS, Moulin TC, Schiöth HB. Sex differences in COVID-19: the role of androgens in disease severity and progression. Endocrine. 2021;71(1):3–8.
Samuel RM, Majd H, Richter MN, Ghazizadeh Z, Zekavat SM, Navickas A, et al. Androgen Signaling regulates SARS-CoV-2 receptor levels and is Associated with severe COVID-19 symptoms in men. Cell Stem Cell. 2020;27(6):876–e88912.
Fitch MN, Phillippi D, Zhang Y, Lucero J, Pandey RS, Liu J, et al. Effects of inhaled air pollution on markers of integrity, inflammation, and microbiota profiles of the intestines in apolipoprotein E knockout mice. Environ Res. 2020;181:108913.
Health impacts [Internet]. [cited 2023 Aug 3]. https://www.who.int/teams/environment-climate-change-and-health/air-quality-and-health/health-impacts
Wu X, Nethery RC, Sabath MB, Braun D, Dominici F. Air pollution and COVID-19 mortality in the United States: strengths and limitations of an ecological regression analysis. Sci Adv. 2020;6(45):eabd4049.
Conticini E, Frediani B, Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ Pollut. 2020;261:114465.
Suwannasual U, Lucero J, Davis G, McDonald JD, Lund AK. Mixed vehicle emissions induces angiotensin II and cerebral microvascular angiotensin receptor expression in C57Bl/6 mice and promotes alterations in Integrity in a blood-brain barrier coculture model. Toxicol Sci. 2019;170(2):525–35.
Md LZ, F W, Jc PRSPS, Jp M et al. P,. Diet-Related Inflammation Is Associated with Worse COVID-19 Outcomes in the UK Biobank Cohort. Nutrients [Internet]. 2023 Feb 9 [cited 2024 Apr 9];15(4). https://pubmed.ncbi.nlm.nih.gov/36839240/
Al Heialy S, Hachim MY, Senok A, Gaudet M, Abou Tayoun A, Hamoudi R, et al. Regulation of angiotensin- converting enzyme 2 in obesity: implications for COVID-19. Front Physiol. 2020;11:555039.
Mathieu E, Escribano-Vazquez U, Descamps D, Cherbuy C, Langella P, Riffault S, et al. Paradigms of lung microbiota functions in Health and Disease, particularly, in Asthma. Front Physiol. 2018;9:1168.
Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. Clemente JC, editor. mBio. 2017;8(1):e02287-16.
Huang YJ, Sethi S, Murphy T, Nariya S, Boushey HA, Lynch SV. Airway Microbiome Dynamics in Exacerbations of Chronic Obstructive Pulmonary Disease. Gilligan PH, editor. J Clin Microbiol. 2014;52(8):2813–23.
Daniel S, Phillippi D, Schneider LJ, Nguyen KN, Mirpuri J, Lund AK. Exposure to diesel exhaust particles results in altered lung microbial profiles, associated with increased reactive oxygen species/reactive nitrogen species and inflammation, in C57Bl/6 wildtype mice on a HF diet. Part Fibre Toxicol. 2021;18(1):3.
Wassenaar TM, Juncos VA, Zimmermann K. Interactions between the gut Microbiome, Lung conditions, and Coronary Heart Disease and how Probiotics affect these. Int J Mol Sci. 2021;22(18):9700.
Li N, He F, Liao B, Zhou Y, Li B, Ran P. Exposure to ambient particulate matter alters the microbial composition and induces immune changes in rat lung. Respir Res. 2017;18(1):143.
Daniel S, Pusadkar V, McDonald J, Mirpuri J, Azad RK, Goven A, et al. Traffic generated emissions alter the lung microbiota by promoting the expansion of Proteobacteria in C57Bl/6 mice placed on a HF diet. Ecotoxicol Environ Saf. 2021;213:112035.
Phillippi DT, Daniel S, Pusadkar V, Youngblood VL, Nguyen KN, Azad RK, et al. Inhaled diesel exhaust particles result in microbiome-related systemic inflammation and altered cardiovascular disease biomarkers in C57Bl/6 male mice. Part Fibre Toxicol. 2022;19(1):10.
Phillippi DT, Daniel S, Nguyen KN, Penaredondo BA, Lund AK. Probiotics function as Immunomodulators in the intestine in C57Bl/6 male mice exposed to inhaled Diesel Exhaust particles on a HF Diet. Cells. 2022;11(9):1445.
Kratzer E, Tian Y, Sarich N, Wu T, Meliton A, Leff A, et al. Oxidative stress contributes to Lung Injury and Barrier Dysfunction via Microtubule Destabilization. Am J Respir Cell Mol Biol. 2012;47(5):688.
Th M. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev [Internet]. 2009 Apr [cited 2024 Apr 9];22(2). https://pubmed.ncbi.nlm.nih.gov/19366914/
Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503.
Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their Molecular Communication with the Immune System. Front Microbiol. 2017;8:2345.
Suwannasual U, Lucero J, McDonald JD, Lund AK. Exposure to traffic-generated air pollutants mediates alterations in brain microvascular integrity in wildtype mice on a HF diet. Environ Res. 2018;160:449–61.
Oppenheim HA, Lucero J, Guyot AC, Herbert LM, McDonald JD, Mabondzo A, et al. Exposure to vehicle emissions results in altered blood brain barrier permeability and expression of matrix metalloproteinases and tight junction proteins in mice. Part Fibre Toxicol. 2013;10(1):62.
Davis G, Lucero J, Fellers C, McDonald JD, Lund AK. The effects of subacute inhaled multi-walled carbon nanotube exposure on signaling pathways associated with cholesterol transport and inflammatory markers in the vasculature of wild-type mice. Toxicol Lett. 2018;296:48–62.
Lund AK, Lucero J, Harman M, Madden MC, McDonald JD, Seagrave JC, et al. The oxidized low-density lipoprotein receptor mediates vascular effects of Inhaled Vehicle emissions. Am J Respir Crit Care Med. 2011;184(1):82–91.
Lund AK, Lucero J, Herbert L, Liu Y, Naik JS. Human immunodeficiency virus transgenic rats exhibit pulmonary hypertension. Am J Physiol-Lung Cell Mol Physiol. 2011;301(3):L315–26.
Lund AK, Lucero J, Lucas S, Madden MC, McDonald JD, Seagrave JC, et al. Vehicular emissions induce vascular MMP-9 expression and activity Associated with Endothelin-1–Mediated pathways. Arterioscler Thromb Vasc Biol. 2009;29(4):511–7.
Acknowledgements
The authors would like to thank Dr. Saskia van Hemert and Winclove Probiotics for their expertise in probiotic formulation and for providing Ecologic Barrier Probiotics for this study. We would also like to thank Dr. Usa Suwannasual and JoAnn Lucero for their assistance with animal exposures and Dr. Imelda Norton at the UNT Vivarium for her support.
Funding
This work was supported by the National Institute of Environmental Health Sciences at the National Institute of Health [2R15ES026795-02] to A.K.L.
Author information
Authors and Affiliations
Contributions
Conceptualization: K.N.-A., S.D., A.K.L.; Methodology: K.N.-A., S.D., D.T.P., A.K.L.; Investigation: K.N.-A., S.D., D.T.P., T.D.A., A.K.L.; Formal analysis: K.N.-A., S.D., D.T.P., T.D.A., B.J., W.I.; Writing (original draft): K.N.-A.; Writing (reviewing and editing): K.N.-A., S.D., D.T.P., T.D.A., A.K.L.; Funding acquisition: A.K.L.; Resources: A.K.L.; Supervision: A.K.L. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experimental procedures were approved by the University of North Texas IACUC under protocol 17009 and followed the Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85 − 23, rev. 1996).
Consent for publication
Not applicable.
Competing interests
The probiotics were provided by Winclove Probiotics and funding from a grant received from the National Institute of Environmental Health Sciences at the National Institute of Health was used to conduct some of the studies described, herein; however, the authors declare no conflict of interest or financial gains to these entities associated with this publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nguyen-Alley, K., Daniel, S., Phillippi, D.T. et al. Diesel exhaust particle inhalation in conjunction with high-fat diet consumption alters the expression of pulmonary SARS-COV-2 infection pathways, which is mitigated by probiotic treatment in C57BL/6 male mice. Part Fibre Toxicol 21, 40 (2024). https://doi.org/10.1186/s12989-024-00601-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12989-024-00601-w